Growth of sessile plants depends on accurate and
timely cellular responses to environmental changes. In the prospect of an
anthropogenic global climate change with expected regional extremes in
temperature and precipitation, it is critical to understand how
plants achieve stress
tolerance
(Link to 'The Copenhagen Diagnosis, 2009').
----------------------------------------
The Ubiquitin Proteasome Pathway
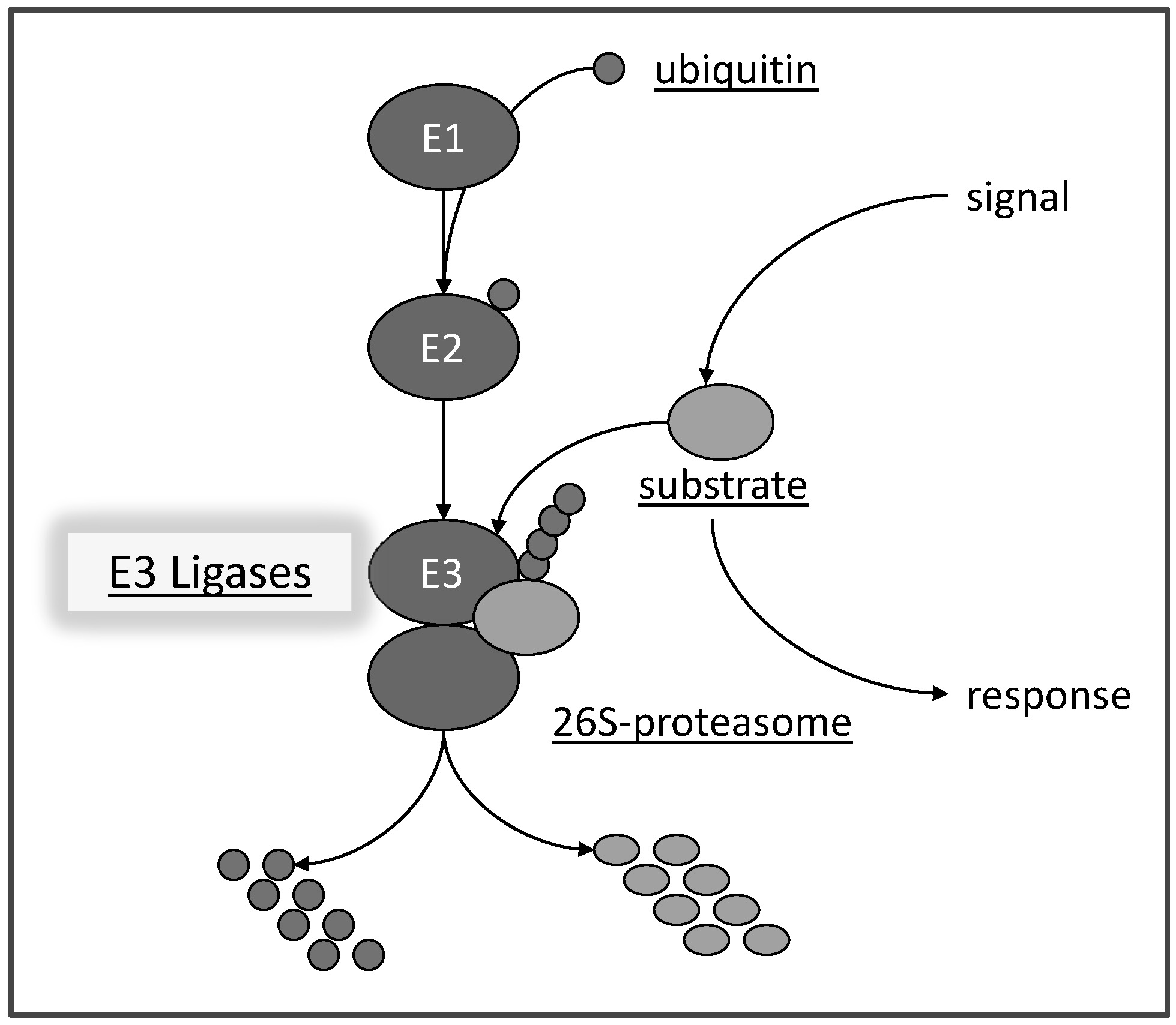
We are interested in resolving aspects of abiotic stress tolerance (salt, drought, heat, etc.) that are facilitated by the ubiquitin proteasome pathway (or ‘kiss of death’ as some people say). The pathway is highly conserved among eukaryotes and affects many processes in plant development and physiology. It requires the concerted activities of several proteins (E1, E2, E3) to activate ubiquitin, a small protein of 79 amino acids, and to build up ubiquitin chains on substrates (Hellmann and Estelle, 2002). This process is referred to as the 'Kiss of Death' since often substrates carrying ubiquitin chains are marked for degradation via a large protein complex, called the 26S proteasome (Fig. 1).
Fig. 1: Scheme of the ubiquitin-proteasome pathway. Ubiquitin is transferred by an ubiquitin activating protein E1 in an ATP-dependent manner to an ubiquitin conjugating E2 protein. E2 can physically assemble with a ubiquitin E3 ligase which facilitate transfer of ubiquitin moieties to a substrate protein. Substrates carrying a ubiquitin chain are a potential target for degradation via the 26S proteasome. The pathway is a central response mediator in eukaryotic cells by degrading proteins or affecting their subcellular location, but potentially is also involved in protein quality control. Note: the pathway is very fast and may degrade proteins within minutes of an initiating signal.
----------------------------------------
DNA Repair
One of the current research topics in the lab is the initiation of DNA repair processes after UV-light exposure. UV-light causes DNA lesions that affect transcriptional processes, and may lead to point mutations if not efficiently repaired in time.
In the last decade it became evident that E3 ubiquitin ligases containing a cullin CUL4, a RING-finger protein RBX1, and the Damaged DNA Binding protein 1 (DDB1) (Bernhardt et al. 2006; Fig. 3A) are key players for recognition of damaged DNA and initiation of repair processes. DDB1 interacts with a multitude of substrate receptors (potentially more than 90 in the plant Arabidopsis thaliana), and thus facilitates ubiquitylation of a broad range of substrate proteins. Not surprisingly, loss of DDB1 is embryo-lethal (Fig. 2) and affects many developmental processes like root and leaf development or flowering (Bernhardt et al. 2010). In addition, some substrate receptors are critical for UV-tolerance, and if they become mutated cause severe UV-hypersensitivity of affected plants (Fig. 3B; Biedermann and Hellmann, 2010).
Fig. 2: Embryo lethal phenotype of an Arabidopsis thaliana ddb1 mutant. Left hand side: siliques with seeds for wild type (upper one) and mutant. Arrows point out aborted seeds. Right hand side: microscopic analysis of seeds at different developmental stages reveals that ddb1 mutants do not progress beyond heart stage in embryo development. Triangles point out embryos.
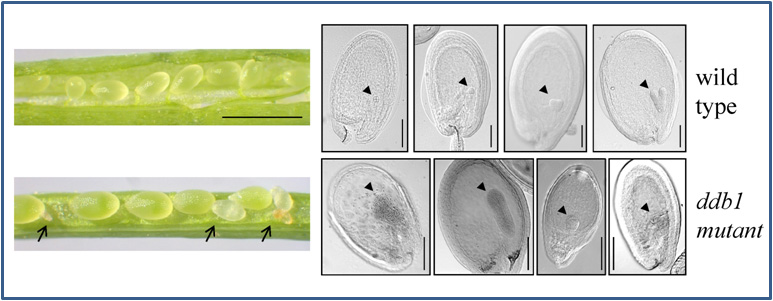
Fig. 3: Critical Pathways for Repair of UV-Induced DNA Lesions in Plants. (A) Plants can repair UV-induced DNA lesions either by the light-dependent activity of photolyases or the light independent pathway of Nucleotide Excision Repair (NER). Our group focuses on mechanisms that activate NER. Critical proteins in this process are a CUL4-based E3 ligase with the core subunits CUL4, RBX1, and DDB1. DDB1 can interact with either CSA (Cockayne Syndrome Factor A; in Arabidopsis thaliana the protein is called ATCSA-1 (Biedermann and Hellmann, 2010)) or DDB2. Both, CSA and DDB2 are key players in damaged DNA recognition and initiating NER. While DDB2 binds directly to damaged DNA and is involved in genome wide control of damaged DNA, CSA does this in concert with CSB (Cockayne Syndrome Factor B; in Arabidopsis called CHR8) and RNA polymerase II, and is only involved in repair of genes actively transcribed. (B) Loss of ATCSA-1 (atcsa-1,1) or DDB2 (ddb2-3) lead to UV-B hypersensitivity of affected plants. Col0, wild type plants; P35S:myc:ATCSA-1, plants with increased ATCSA-1 levels (note: increased ATCSA-1 does not lead to increased UV-B tolerance). 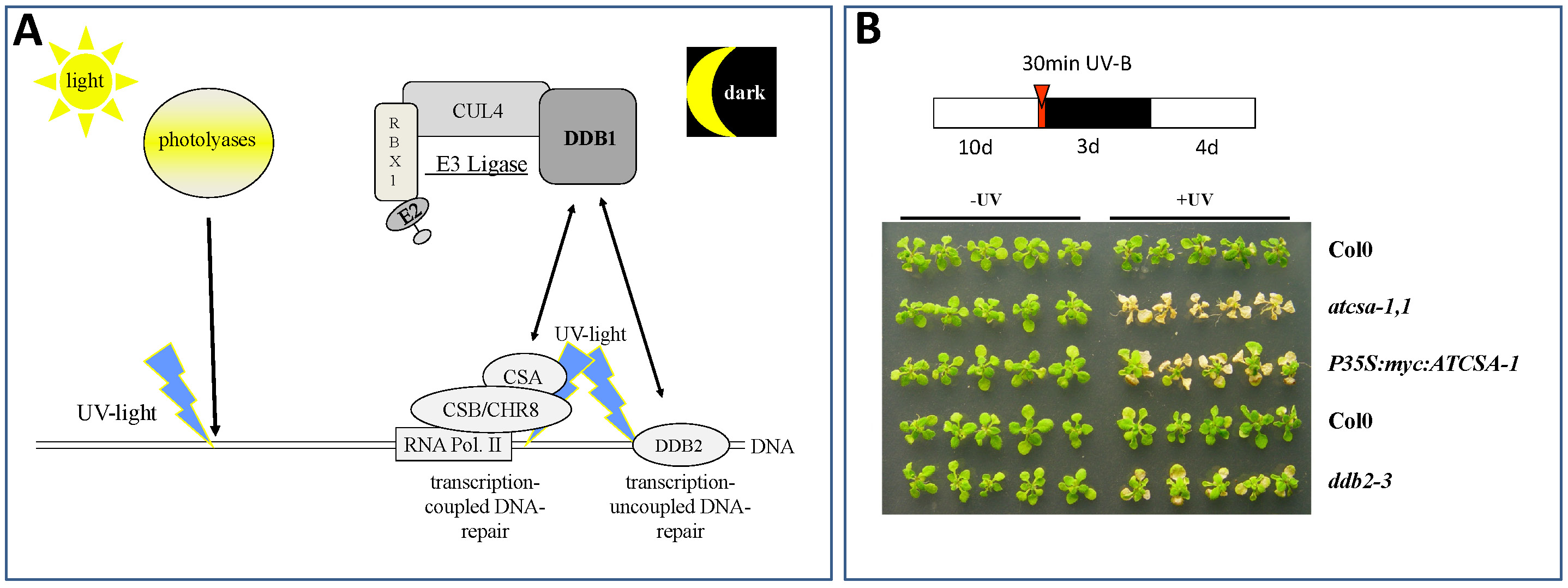
Transcriptional Control
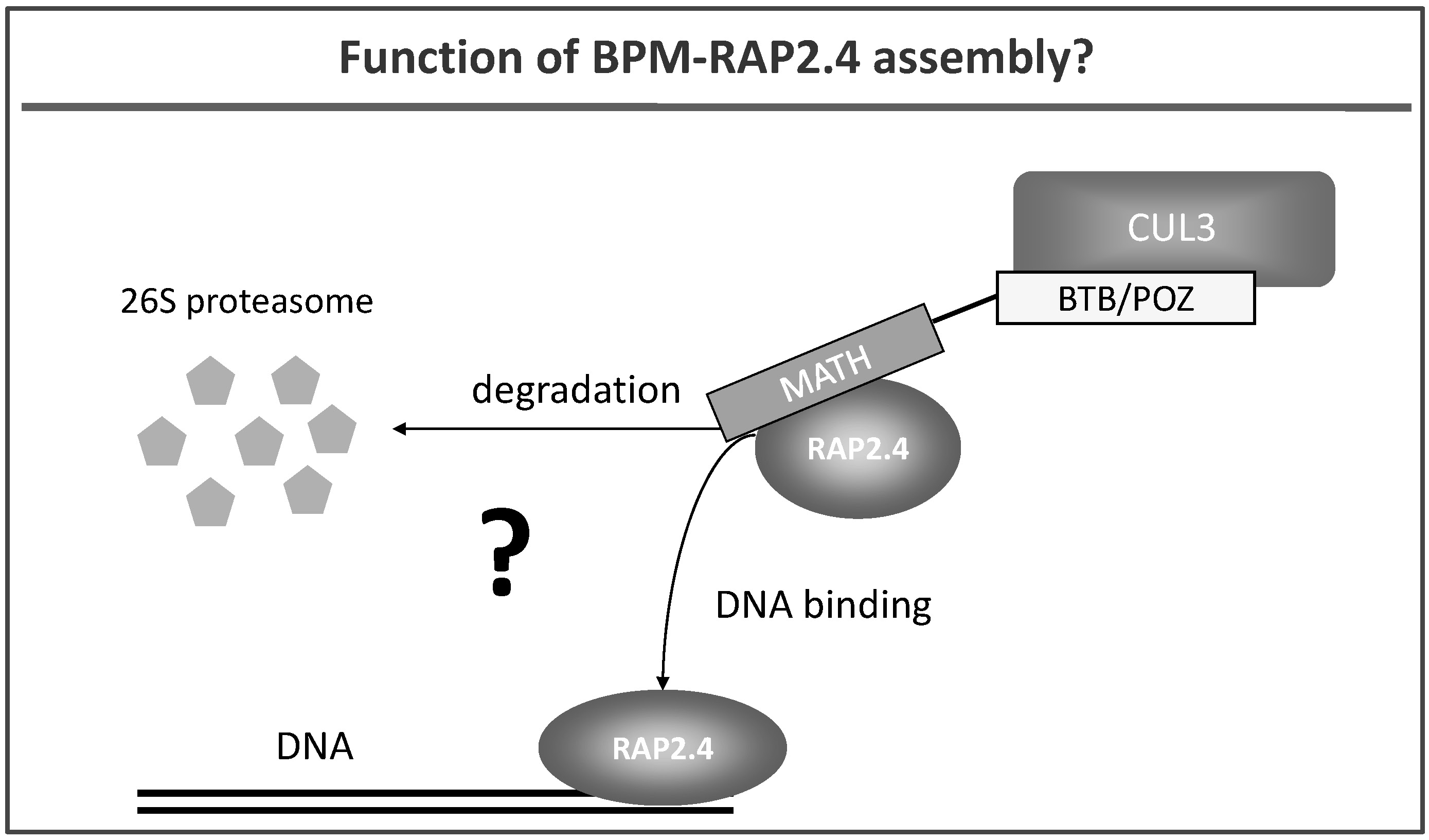
We also work on E3 ubiquitin ligases that contain a cullin CUL3. CUL3 proteins are able to interact with a group of proteins called BTB/POZ-MATH (BPM) (Weber et al. 2005) (Fig. 4). BPM proteins are discussed to function as substrate receptors in plants but functional proof is basically missing. We have identified transcription factors (e.g. RAP2.4; Fig. 4) that play a role in abiotic stress tolerance as potential substrate proteins for CUL3-based E3 ligases (Weber and Hellmann, 2009), and are currently resolving the impact of the CUL3 complex on transcriptional processes and stress tolerance in plants.
Fig. 4: Hypothetical roles of CUL3 and BTB-POZ/MATH (BPM) proteins: they are likely to either target the transcription factor RAP2.4 for proteolytic degradation via the 26S proteasome or BPM proteins bind to RAP2.4 without initiating its proteolysis. Here BPM-RAP2.4 assembly might alter, for example, the DNA binding ability of the transcription factor.
----------------------------------------
VITAMIN B6
Independently of the ubiquitin proteasome pathway, we are also interested in vitamin B6 metabolism. Besides being a critical cofactor for more than 140 biochemical reactions, the vitamin is also considered to be a potent antioxidant, and to have many beneficial impacts on human health (for an overview see Hellmann and Mooney, 2010).
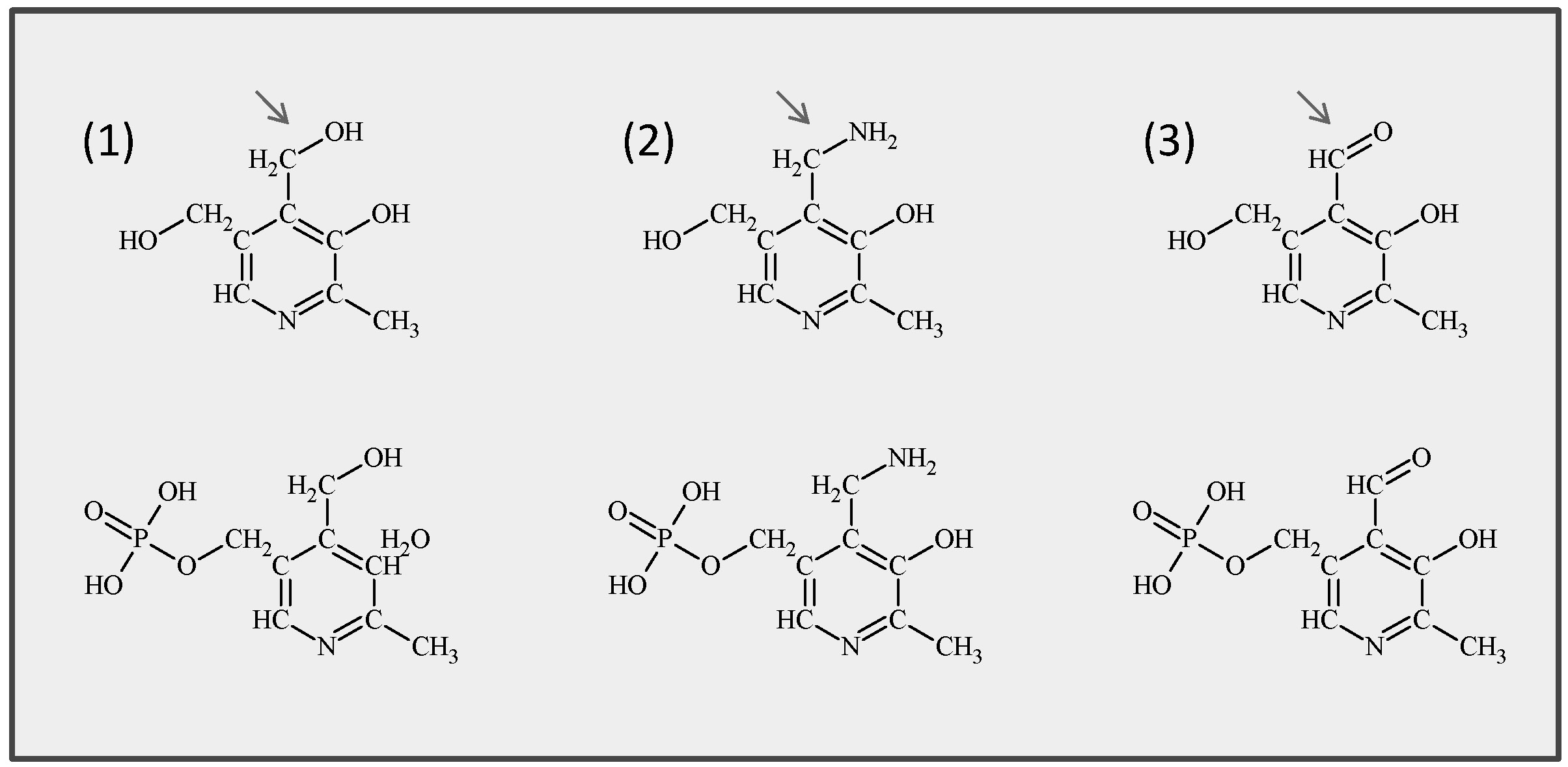
Vitamin B6 comprises a group of six derivatives: pyridoxine (PN), pyridoxamine (PM), and pyridoxal (PL) and their phosphorylated forms which function as the actual cofactors (Fig. 5).
Fig. 5: Chemical structures of the six common B6 vitamers. (1) Pyridoxine and its phosphorylated form pyridoxine 5'-phosphate (PLP), (2) pyridoxamine and pyridoxamine 5'-phosphate, (3) pyridoxal and pyridoxal 5'-phosphate. Arrows indicate the variable group at the 4’ position.
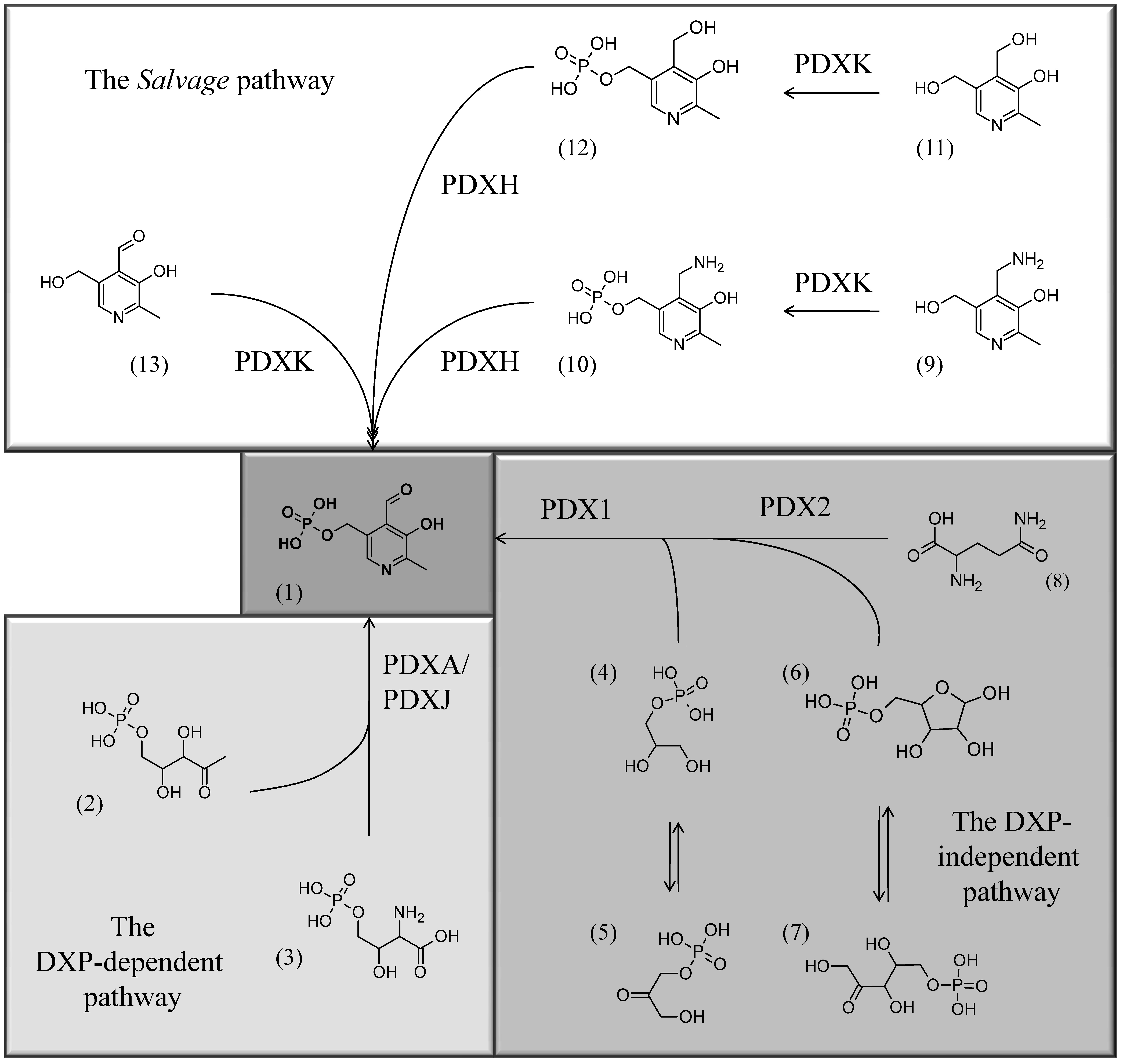
De novo biosynthesis of vitamin B6 (or actually pyridoxal-5-phosphate (PLP) in plants and fungi) is catalyzed by two proteins, PDX1 and PDX2, which together form a PLP synthase complex. Plants lacking either PDX1 or PDX2 die if not rescued by external supply of the vitamin.
Fig.6: The three known pathways for PLP biosynthesis: one salvage pathway, and two de novo pathways, a Deoxyxylulose (DXP)-dependent one and a DXP-independent one (for more information see Mooney and Hellmann, 2010). Chemical structures: (1) PLP, (2) Deoxyxylulose 5’-phosphate, (3) 4-(Phosphohydroxy)-L-threonine, (4) Glyceraldehyde 3’-phosphate, (5) Dihydroxyacetone phosphate, (6) Ribose 5’-phosphate, (7) Ribulose 5’-phosphate, (8) glutamine, (9) PM, (10) PMP, (11) PN, (12) PNP, (13) PL.
In addition, the vitamin has been brought into context with different developmental processes (root and leaf development, flowering (Wagner et al. 2006; Fig. 7), and abiotic stress tolerance (plants with reduced vitamin B6 are light sensitive, and show increased sensitivity towards UV-B, salt and osmotic stress (Chen and Xiong, 2005; Denslow et al. 2007; Titiz et al. 2006).
Our current research focuses on regulatory aspect of vitamin B6 biosynthesis, its impact on plant development and why the vitamin is required for stress tolerance.
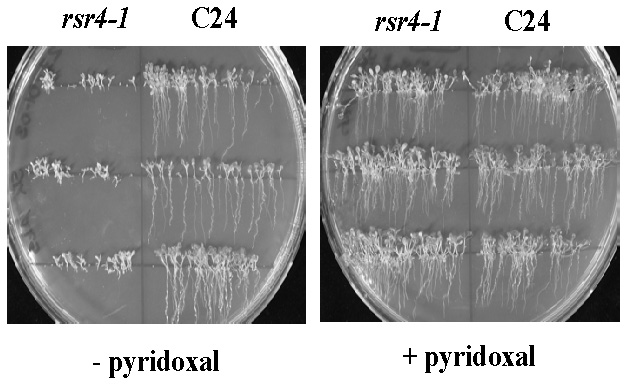
Fig. 7: Normalization of root growth in the vitamin B6 biosynthesis mutant rsr4-1. rsr4-1 is affected in a PDX1 gene which causes a nearly 70% reduction in vitamin B6 content. Most likely as a consequence of reduced vitamin B6, root development is strongly disturbed (left hand site). Note: external supply of pyridoxal can rescue this phenotype and generates plants indistinguishable from wild type C24 (right hand site).